Reading and Working with Tabular Data
Overview
Teaching: 50 min
Exercises: 40 minQuestions
What is a Pandas data frame?
How do I get an overview on my tabular data?
How do I read tabular data in different formats?
How do I access subsets of a data frame?
How do I calculate simple statistics like the mean?
Objectives
Read data in various formats.
Select individual values and subsections from data.
Transform a data frame from messy to tidy data.
Understand common data workflows (ETL) and strategies (split-apply-combine).
Read data
We use Pandas for reading, writing, and handling tabular data in Python as it is the de facto standard tool to do so.
Just as np is by convention the alias for numpy, so is pd for pandas.
import pandas as pd
We start by reading some data from a comma-separated values (CSV) file.
growth = pd.read_csv("data/yeast-growth.csv")
Inspect your data
Depending on your current Python environment a pandas.DataFrame object will automatically be displayed as text formatted to a table or as a prettier HTML table.
growth
well timepoint od concentration_level concentration
0 a 1 0.017 low 0.01
1 b 1 0.017 low 0.03
2 c 1 0.018 medium 1.00
3 d 1 0.017 medium 3.00
4 e 1 0.017 medium 30.00
.. ... ... ... ... ...
450 c 65 0.228 medium 1.00
451 d 65 0.221 medium 3.00
452 e 65 0.065 medium 30.00
453 f 65 0.035 high 100.00
454 g 65 0.031 high 300.00
[455 rows x 5 columns]
By default, the output is configured to not show too many rows and columns. We can access more specific parts of a data frame using specific methods.
The beginning of a data frame can be accessed with the head method.
growth.head()
well timepoint od concentration_level concentration
0 a 1 0.017 low 0.01
1 b 1 0.017 low 0.03
2 c 1 0.018 medium 1.00
3 d 1 0.017 medium 3.00
4 e 1 0.017 medium 30.00
Correspondingly, the end of a data frame can be accessed with the tail method.
growth.tail()
well timepoint od concentration_level concentration
450 c 65 0.228 medium 1.0
451 d 65 0.221 medium 3.0
452 e 65 0.065 medium 30.0
453 f 65 0.035 high 100.0
454 g 65 0.031 high 300.0
You can also ask for a random number of rows. This is great for inspection since you have a chance to see rows from different parts of the data frame. It can also be convenient to develop an analysis with a subset of your data first because it will be faster (this matters if you work with tables that contain millions of rows).
growth.sample(10)
well timepoint od concentration_level concentration
332 d 48 0.120 medium 3.00
263 e 38 0.046 medium 30.00
139 g 20 0.032 high 300.00
376 f 54 0.036 high 100.00
415 c 60 0.231 medium 1.00
424 e 61 0.063 medium 30.00
392 a 57 0.234 low 0.01
142 c 21 0.049 medium 1.00
312 e 45 0.051 medium 30.00
12 f 2 0.015 high 100.00
Notice the random indices.
Summaries
Looking at the actual values in your data is important to develop an understanding of them if you work with new data. However, in order to get an overview on your data, there are some useful summary methods.
If you just want to know the dimensions of your data frame, you can do so with the shape attribute.
growth.shape
(455, 5)
As you will see later on, it is sometimes necessary to look at the data types that pandas has inferred from your table.
growth.dtypes
well object
timepoint int64
od float64
concentration_level object
concentration float64
dtype: object
The info method conveniently combines information on the shape and types in your data frame. In addition, it tells you about the index used and the memory consumption.
growth.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 455 entries, 0 to 454
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 well 455 non-null object
1 timepoint 455 non-null int64
2 od 455 non-null float64
3 concentration_level 455 non-null object
4 concentration 455 non-null float64
dtypes: float64(2), int64(1), object(2)
memory usage: 17.9+ KB
Instead of summarizing the data frame object itself, you can get a high-level overview on the statistical distribution of your data.
growth.describe()
timepoint od concentration
count 455.000000 455.000000 455.000000
mean 33.000000 0.077193 62.005714
std 18.782314 0.068110 102.928567
min 1.000000 0.015000 0.010000
25% 17.000000 0.032000 0.030000
50% 33.000000 0.042000 3.000000
75% 49.000000 0.099500 100.000000
max 65.000000 0.237000 300.000000
By default, this will only describe numeric columns. You can also get an overview on all columns including text and categorical columns (generally of type object).
growth.describe(include="all")
well timepoint od concentration_level concentration
count 455 455.000000 455.000000 455 455.000000
unique 7 NaN NaN 3 NaN
top a NaN NaN medium NaN
freq 65 NaN NaN 195 NaN
mean NaN 33.000000 0.077193 NaN 62.005714
std NaN 18.782314 0.068110 NaN 102.928567
min NaN 1.000000 0.015000 NaN 0.010000
25% NaN 17.000000 0.032000 NaN 0.030000
50% NaN 33.000000 0.042000 NaN 3.000000
75% NaN 49.000000 0.099500 NaN 100.000000
max NaN 65.000000 0.237000 NaN 300.000000
The NaN values displayed in the output are simply Pandas’ concept of missing values. Just pretend that they are empty cells.
Wording
I try to use data frame when talking about the object for computation and table when talking about data more generally; but I might mix them up as in my mind they are the same concept.
Tangent: Tidy Data
Hadley Wickham coined the concept of tidy data for statistics. What is tidy data?
You can also dive into the original publication from 2014 but it suffices to say that a tidy data structure is more intuitive for data visualization (plotting) and statistical analysis.
Reading files from different locales
Depending on what country’s standard your computer is set to (the ‘locale’), software such as Excel will use different characters to separate fields, for example, the default for a computer with the DK locale will be to use
;to separate fields and,to separate decimals. Try finding the right arguments topandas.read_tableto get something sensible out of"data/example-uk.txt"and"data/example-dk.txt". Use the inspection methods from before to see how pandas reads the data.UK
Solution
uk_data = pd.read_table("data/example-uk.txt") uk_dataname;height;age;income 0 Ian;183;27;12,000.01 1 Peter;162;28;11,000.50 2 Bernhard;173;30;10,000.00 3 Steven;163;32;12,500.00uk_data = pd.read_table("data/example-uk.txt", sep=";") uk_dataname height age income 0 Ian 183 27 12,000.01 1 Peter 162 28 11,000.50 2 Bernhard 173 30 10,000.00 3 Steven 163 32 12,500.00uk_data.dtypesname object height int64 age int64 income object dtype: objectuk_data = pd.read_table("data/example-uk.txt", sep=";", thousands=",") uk_dataname height age income 0 Ian 183 27 12000.01 1 Peter 162 28 11000.50 2 Bernhard 173 30 10000.00 3 Steven 163 32 12500.00uk_data.dtypesname object height int64 age int64 income float64 dtype: objectDK
Solution
dk_data = pd.read_table("data/example-dk.txt") dk_dataname;height;age;income 0 Ian;183;27;12000,01 1 Peter;162;28;11100,50 2 Bernhard;173;30;11000,00 3 Steven;163;32;12500,00dk_data = pd.read_table("data/example-dk.txt", sep=";") dk_dataname height age income 0 Ian 183 27 12000,01 1 Peter 162 28 11100,50 2 Bernhard 173 30 11000,00 3 Steven 163 32 12500,00dk_data.dtypesname object height int64 age int64 income object dtype: objectdk_data = pd.read_table("data/example-dk.txt", sep=";", decimal=",") dk_dataname height age income 0 Ian 183 27 12000.01 1 Peter 162 28 11100.50 2 Bernhard 173 30 11000.00 3 Steven 163 32 12500.00dk_data.dtypesname object height int64 age int64 income float64 dtype: object
Other data formats
Excel
When you read data from spreadsheets (.xls, .xlsx, .ods), you do not need to worry about field separators or number formatting (but you need extra packages such as xlrd, openpyxl, or odfpy).
excel_data = pd.read_excel("data/example-dk.xlsx")
excel_data
name height age income
0 Ian 183 27 12000.01
1 Peter 162 28 11100.50
2 Bernhard 173 30 11000.00
3 Steven 163 32 12500.00
However, you may need to be careful with Excel’s interpretation of strings as dates (see Gene name errors are widespread in the scientific literature and Guidelines for human gene nomenclature) when loading tabular files in Excel.
Pandas will try to detect the right dependency for opening a spreadsheet.
date_data = pd.read_excel("data/example-datetime.ods")
date_data
date time
0 2020-03-02 03:34:23
1 2021-02-02 02:24:00
2 2021-09-01 16:01:22
Common date and time formats are automatically detected and converted to suitable Python data types.
date_data.dtypes
date datetime64[ns]
time object
dtype: object
You can conveniently combine multiple columns into one datetime object.
pd.read_excel("data/example-datetime.ods", parse_dates={"datetime": ["date", "time"]})
datetime
0 2020-03-02 03:34:23
1 2021-02-02 02:24:00
2 2021-09-01 16:01:22
Further formats
There are many more formats that Pandas can read data from. Just type pd.read_
and press tab to see the list of auto-completion options. A few notable ones:
- All file-based read functions accept a remote URL as an option. We could have loaded the growth data with
pd.read_csv(
"https://raw.githubusercontent.com/data-science-for-biotech/python-pandas-viz-ml/gh-pages/_episodes_rmd/data/yeast-growth.csv"
)
well timepoint od concentration_level concentration
0 a 1 0.017 low 0.01
1 b 1 0.017 low 0.03
2 c 1 0.018 medium 1.00
3 d 1 0.017 medium 3.00
4 e 1 0.017 medium 30.00
.. ... ... ... ... ...
450 c 65 0.228 medium 1.00
451 d 65 0.221 medium 3.00
452 e 65 0.065 medium 30.00
453 f 65 0.035 high 100.00
454 g 65 0.031 high 300.00
[455 rows x 5 columns]
- In the context of the web,
read_jsoncan be very handy - Tables are often stored in SQL databases, in that case
read_sqlis your friend
Accessing data
Let’s look at our old friend the growth data table again.
growth
well timepoint od concentration_level concentration
0 a 1 0.017 low 0.01
1 b 1 0.017 low 0.03
2 c 1 0.018 medium 1.00
3 d 1 0.017 medium 3.00
4 e 1 0.017 medium 30.00
.. ... ... ... ... ...
450 c 65 0.228 medium 1.00
451 d 65 0.221 medium 3.00
452 e 65 0.065 medium 30.00
453 f 65 0.035 high 100.00
454 g 65 0.031 high 300.00
[455 rows x 5 columns]
By label
We can access individual columns (pandas.Series) by their names as string.
growth["od"]
0 0.017
1 0.017
2 0.018
3 0.017
4 0.017
...
450 0.228
451 0.221
452 0.065
453 0.035
454 0.031
Name: od, Length: 455, dtype: float64
Notice how the display of series is different from data frames.
We can make powerful label based selections with the loc attribute.
growth.loc[:, "od"]
0 0.017
1 0.017
2 0.018
3 0.017
4 0.017
...
450 0.228
451 0.221
452 0.065
453 0.035
454 0.031
Name: od, Length: 455, dtype: float64
Our data frames have two dimensions: the rows and columns. Here, we specify that
we want all rows : and a single column "od". We can also select one or more
columns as a list which will always return a data frame.
growth.loc[:, ["od"]]
od
0 0.017
1 0.017
2 0.018
3 0.017
4 0.017
.. ...
450 0.228
451 0.221
452 0.065
453 0.035
454 0.031
[455 rows x 1 columns]
We use at to access an individual element with labels.
growth.at[451, "od"]
0.221
By index
We might be tempted to access a column by its index position directly.
growth[2]
Error in py_call_impl(callable, dots$args, dots$keywords): KeyError: 2
Detailed traceback:
File "<string>", line 1, in <module>
File "/home/moritz/.pyenv/versions/3.8.10/envs/biotech/lib/python3.8/site-packages/pandas/core/frame.py", line 3024, in __getitem__
indexer = self.columns.get_loc(key)
File "/home/moritz/.pyenv/versions/3.8.10/envs/biotech/lib/python3.8/site-packages/pandas/core/indexes/base.py", line 3082, in get_loc
raise KeyError(key) from err
This does not work. Only labels are allowed here. Instead we use iloc for
index location.
growth.iloc[:, 2]
0 0.017
1 0.017
2 0.018
3 0.017
4 0.017
...
450 0.228
451 0.221
452 0.065
453 0.035
454 0.031
Name: od, Length: 455, dtype: float64
Again, we can select one or more columns as a data frame.
growth.iloc[:, [2]]
od
0 0.017
1 0.017
2 0.018
3 0.017
4 0.017
.. ...
450 0.228
451 0.221
452 0.065
453 0.035
454 0.031
[455 rows x 1 columns]
We can also access individual elements.
growth.iat[451, 2]
0.221
By slices
We can also define label-based ranges over rows and columns.
growth.loc[:, "od":"concentration"]
od concentration_level concentration
0 0.017 low 0.01
1 0.017 low 0.03
2 0.018 medium 1.00
3 0.017 medium 3.00
4 0.017 medium 30.00
.. ... ... ...
450 0.228 medium 1.00
451 0.221 medium 3.00
452 0.065 medium 30.00
453 0.035 high 100.00
454 0.031 high 300.00
[455 rows x 3 columns]
growth.loc[10:15, :]
well timepoint od concentration_level concentration
10 d 2 0.015 medium 3.00
11 e 2 0.019 medium 30.00
12 f 2 0.015 high 100.00
13 g 2 0.016 high 300.00
14 a 3 0.016 low 0.01
15 b 3 0.015 low 0.03
locBe careful that in label-based accession with
loc, ranges are inclusive unlike numeric ranges in Python in general. Also be aware that since we have an integer index here, the label coincides with the index position.
growth.iloc[10:15, :]
well timepoint od concentration_level concentration
10 d 2 0.015 medium 3.00
11 e 2 0.019 medium 30.00
12 f 2 0.015 high 100.00
13 g 2 0.016 high 300.00
14 a 3 0.016 low 0.01
N.B.: Index location ranges are exclusive.
By conditions
Pandas inherits from numpy the conditional selection of elements. These are essentially Boolean series or data frames known as masks.
growth[growth["od"] > 0.2]
well timepoint od concentration_level concentration
308 a 45 0.201 low 0.01
315 a 46 0.208 low 0.01
322 a 47 0.217 low 0.01
329 a 48 0.227 low 0.01
330 b 48 0.205 low 0.03
336 a 49 0.229 low 0.01
337 b 49 0.213 low 0.03
343 a 50 0.232 low 0.01
344 b 50 0.219 low 0.03
350 a 51 0.233 low 0.01
351 b 51 0.224 low 0.03
357 a 52 0.237 low 0.01
358 b 52 0.229 low 0.03
364 a 53 0.236 low 0.01
365 b 53 0.230 low 0.03
366 c 53 0.207 medium 1.00
371 a 54 0.235 low 0.01
372 b 54 0.231 low 0.03
373 c 54 0.213 medium 1.00
378 a 55 0.237 low 0.01
379 b 55 0.230 low 0.03
380 c 55 0.218 medium 1.00
385 a 56 0.232 low 0.01
386 b 56 0.231 low 0.03
387 c 56 0.226 medium 1.00
392 a 57 0.234 low 0.01
393 b 57 0.233 low 0.03
394 c 57 0.230 medium 1.00
399 a 58 0.232 low 0.01
400 b 58 0.228 low 0.03
401 c 58 0.231 medium 1.00
406 a 59 0.230 low 0.01
407 b 59 0.228 low 0.03
408 c 59 0.230 medium 1.00
413 a 60 0.228 low 0.01
414 b 60 0.225 low 0.03
415 c 60 0.231 medium 1.00
420 a 61 0.227 low 0.01
421 b 61 0.225 low 0.03
422 c 61 0.230 medium 1.00
427 a 62 0.226 low 0.01
428 b 62 0.222 low 0.03
429 c 62 0.230 medium 1.00
430 d 62 0.205 medium 3.00
434 a 63 0.229 low 0.01
435 b 63 0.225 low 0.03
436 c 63 0.230 medium 1.00
437 d 63 0.212 medium 3.00
441 a 64 0.224 low 0.01
442 b 64 0.221 low 0.03
443 c 64 0.231 medium 1.00
444 d 64 0.217 medium 3.00
448 a 65 0.224 low 0.01
449 b 65 0.220 low 0.03
450 c 65 0.228 medium 1.00
451 d 65 0.221 medium 3.00
These masks can be combined using Boolean logic, making them very powerful tools for selecting just the right data.
growth[(growth["od"] > 0.2) & (growth["concentration_level"] == "medium")]
well timepoint od concentration_level concentration
366 c 53 0.207 medium 1.0
373 c 54 0.213 medium 1.0
380 c 55 0.218 medium 1.0
387 c 56 0.226 medium 1.0
394 c 57 0.230 medium 1.0
401 c 58 0.231 medium 1.0
408 c 59 0.230 medium 1.0
415 c 60 0.231 medium 1.0
422 c 61 0.230 medium 1.0
429 c 62 0.230 medium 1.0
430 d 62 0.205 medium 3.0
436 c 63 0.230 medium 1.0
437 d 63 0.212 medium 3.0
443 c 64 0.231 medium 1.0
444 d 64 0.217 medium 3.0
450 c 65 0.228 medium 1.0
451 d 65 0.221 medium 3.0
You can combine masks with loc to also sub-select columns.
growth.loc[(growth["od"] > 0.2) & (growth["concentration_level"] == "medium"), ["od", "timepoint"]]
od timepoint
366 0.207 53
373 0.213 54
380 0.218 55
387 0.226 56
394 0.230 57
401 0.231 58
408 0.230 59
415 0.231 60
422 0.230 61
429 0.230 62
430 0.205 62
436 0.230 63
437 0.212 63
443 0.231 64
444 0.217 64
450 0.228 65
451 0.221 65
Sometimes these conditions can be tedious to write, use the query method for a
shorter form. Be careful, query uses Python’s condition logic, not numpy’s.
growth.query("od > 0.2 and concentration_level == 'medium'")
well timepoint od concentration_level concentration
366 c 53 0.207 medium 1.0
373 c 54 0.213 medium 1.0
380 c 55 0.218 medium 1.0
387 c 56 0.226 medium 1.0
394 c 57 0.230 medium 1.0
401 c 58 0.231 medium 1.0
408 c 59 0.230 medium 1.0
415 c 60 0.231 medium 1.0
422 c 61 0.230 medium 1.0
429 c 62 0.230 medium 1.0
430 d 62 0.205 medium 3.0
436 c 63 0.230 medium 1.0
437 d 63 0.212 medium 3.0
443 c 64 0.231 medium 1.0
444 d 64 0.217 medium 3.0
450 c 65 0.228 medium 1.0
451 d 65 0.221 medium 3.0
query is fancy enough to know how to use variables.
threshold = 0.2
growth.query("od > @threshold and concentration_level == 'medium'")
well timepoint od concentration_level concentration
366 c 53 0.207 medium 1.0
373 c 54 0.213 medium 1.0
380 c 55 0.218 medium 1.0
387 c 56 0.226 medium 1.0
394 c 57 0.230 medium 1.0
401 c 58 0.231 medium 1.0
408 c 59 0.230 medium 1.0
415 c 60 0.231 medium 1.0
422 c 61 0.230 medium 1.0
429 c 62 0.230 medium 1.0
430 d 62 0.205 medium 3.0
436 c 63 0.230 medium 1.0
437 d 63 0.212 medium 3.0
443 c 64 0.231 medium 1.0
444 d 64 0.217 medium 3.0
450 c 65 0.228 medium 1.0
451 d 65 0.221 medium 3.0
Views
Selecting a subset of your data frame by any of the previous means creates a view on that part of the data frame. This is faster since the data is not copied and lets you change elements in a part of the table. Be careful that this may lead to unintended consequences.
growth.loc[0:5, :] = None
growth.head(10)
well timepoint od concentration_level concentration
0 None NaN NaN None NaN
1 None NaN NaN None NaN
2 None NaN NaN None NaN
3 None NaN NaN None NaN
4 None NaN NaN None NaN
5 None NaN NaN None NaN
6 g 1.0 0.015 high 300.00
7 a 2.0 0.015 low 0.01
8 b 2.0 0.018 low 0.03
9 c 2.0 0.021 medium 1.00
A comprehensive tutorial can be found in the Pandas documentation.
Working with data
Just like numpy, Pandas offers basic statistical methods out-of-the-box. We can apply them to single columns,
growth["od"].min()
0.015
multiple columns,
growth[["od", "concentration"]].max()
od 0.237
concentration 300.000
dtype: float64
or even rows.
growth[["od", "concentration"]].mean(axis=1)
0 NaN
1 NaN
2 NaN
3 NaN
4 NaN
...
450 0.6140
451 1.6105
452 15.0325
453 50.0175
454 150.0155
Length: 455, dtype: float64
growth[["od", "concentration"]].std(axis=1)
0 NaN
1 NaN
2 NaN
3 NaN
4 NaN
...
450 0.545886
451 1.965050
452 21.167241
453 70.685929
454 212.110114
Length: 455, dtype: float64
Split-apply-combine
Another powerful operation and a very common strategy for data transformation is
split, apply,
combine.
This pattern is provided by pandas in the form of the groupby method.
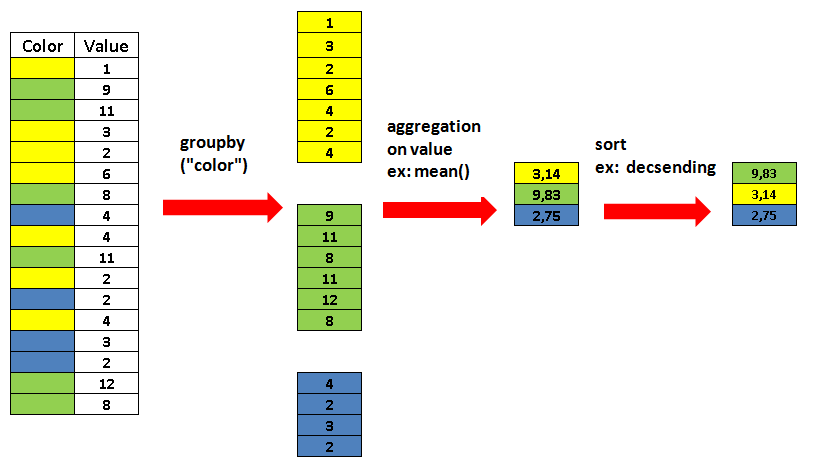
growth.groupby("well")[["od", "concentration"]].mean()
od concentration
well
a 0.123500 0.01
b 0.117969 0.03
c 0.109375 1.00
d 0.088328 3.00
e 0.043891 30.00
f 0.033937 100.00
g 0.029738 300.00
Cleaning messy data
Can you transform this messy table into a tidy one? You may need the
meltfunction to change the table layout.messy = pd.read_csv("data/yeast-growth-messy.csv")Solution
messyV1 V2 V3 V4 V5 V6 ... V63 V64 V65 V66 V67 V68 0 Test1 low 0.01 0.017 0.015 0.016 ... 0.228 0.227 0.226 0.229 0.224 0.224 1 Test1 low 0.03 0.017 0.018 0.015 ... 0.225 0.225 0.222 0.225 0.221 0.220 2 Test1 medium 1.00 0.018 0.021 0.016 ... 0.231 0.230 0.230 0.230 0.231 0.228 3 Test1 medium 3.00 0.017 0.015 0.017 ... 0.190 0.196 0.205 0.212 0.217 0.221 4 Test1 medium 30.00 0.017 0.019 0.016 ... 0.062 0.063 0.062 0.063 0.063 0.065 5 Test1 high 100.00 0.016 0.015 0.015 ... 0.037 0.035 0.035 0.034 0.035 0.035 6 Test1 high 300.00 0.015 0.016 0.015 ... 0.031 0.032 0.031 0.031 0.030 0.031 [7 rows x 68 columns]messy.columnsIndex(['V1', 'V2', 'V3', 'V4', 'V5', 'V6', 'V7', 'V8', 'V9', 'V10', 'V11', 'V12', 'V13', 'V14', 'V15', 'V16', 'V17', 'V18', 'V19', 'V20', 'V21', 'V22', 'V23', 'V24', 'V25', 'V26', 'V27', 'V28', 'V29', 'V30', 'V31', 'V32', 'V33', 'V34', 'V35', 'V36', 'V37', 'V38', 'V39', 'V40', 'V41', 'V42', 'V43', 'V44', 'V45', 'V46', 'V47', 'V48', 'V49', 'V50', 'V51', 'V52', 'V53', 'V54', 'V55', 'V56', 'V57', 'V58', 'V59', 'V60', 'V61', 'V62', 'V63', 'V64', 'V65', 'V66', 'V67', 'V68'], dtype='object')del messy["V1"]from string import ascii_lowercasemessy["well"] = list(ascii_lowercase[:len(messy)])long_data = pd.melt(messy, id_vars=["V2", "V3", "well"]) long_dataV2 V3 well variable value 0 low 0.01 a V4 0.017 1 low 0.03 b V4 0.017 2 medium 1.00 c V4 0.018 3 medium 3.00 d V4 0.017 4 medium 30.00 e V4 0.017 .. ... ... ... ... ... 450 medium 1.00 c V68 0.228 451 medium 3.00 d V68 0.221 452 medium 30.00 e V68 0.065 453 high 100.00 f V68 0.035 454 high 300.00 g V68 0.031 [455 rows x 5 columns]long_data["variable"].replace(dict(zip(long_data["variable"].unique(), range(1, len(long_data["variable"].unique()) + 1))), inplace=True)long_data.rename(columns={"V2": "concentration_level", "V3": "concentration", "variable": "timepoint"}, inplace=True)long_dataconcentration_level concentration well timepoint value 0 low 0.01 a 1 0.017 1 low 0.03 b 1 0.017 2 medium 1.00 c 1 0.018 3 medium 3.00 d 1 0.017 4 medium 30.00 e 1 0.017 .. ... ... ... ... ... 450 medium 1.00 c 65 0.228 451 medium 3.00 d 65 0.221 452 medium 30.00 e 65 0.065 453 high 100.00 f 65 0.035 454 high 300.00 g 65 0.031 [455 rows x 5 columns]
Tangent: Extract, Transform, Load (ETL)
The process we have just seen is known as extract, transform, load or simply ETL. It describes the ubiquitous data workflow of reading data (extract with, for example, pd.read_csv), cleaning up the data (transform) as we have done with the messy growth curves above, and storing the tidy data again for subsequent use (load with, for example, to_excel).
Outlook
- Tidy pandas data frames are excellent structures for visualizing data, either with the built-in methods or other packages. This will be part of the next episode.
- The package statsmodels can be used to fit statistical models to tidy data frames in a convenient fashion using
R-style formulas. - Data frames due to their column names are more descriptive than pure numeric matrices (
numpy.ndarray) which is handy in many tasks, for example, in machine learning as shown in the last episode.
Key Points
Use
pandas.read_*andpandas.DataFrame.to_*to import / export data.The method
infodescribes the data frame object.The method
describesummarizes value distributions in columns.You can use labels or index locations to select both subsets and elements from your data frame.
Selection using conditions is very powerful.
Selections create views on your original data.
Use
mean,max,min, and others to calculate simple statistics.Use split-apply-combine to calculate statistics within groups in a data frame.